Introduction:
In the rapidly evolving domains of physics, chemistry, materials science, and atomistic simulations, a multitude of datasets have been curated to drive innovation and exploration. These datasets encompass both experimental and computational data, enabling researchers to conduct high-throughput screenings and seamlessly integrate machine learning (ML) techniques. They play a crucial role in advancing big data initiatives and harnessing artificial intelligence (AI) to predict and design new materials. Below is an extensive collection of key datasets that serve as invaluable resources for researchers and scientists engaged in these interdisciplinary fields.
Motivation:
Datasets are vital in materials science because they provide structured, high-quality data that fuel computational models, validate experiments, and accelerate the discovery of novel materials. By offering pre-curated collections of properties like crystal structures, electronic band gaps, or reaction energies, these datasets eliminate the need for time-intensive data generation, enabling researchers to focus on analysis and innovation. They empower AI and ML to uncover patterns and predict material behaviors, driving advancements in fields like energy storage, catalysis, and nanotechnology, while fostering reproducible and collaborative research.
Purpose:
This table compiles key open-access, free to use and community-curated datasets to guide researchers in selecting data resources that align with their scientific objectives, whether for atomistic simulations, machine learning model training, or high-throughput materials screening. It provides a concise overview of each dataset’s scope, data types, size, and tools, enabling users to compare options and leverage these collections for cutting-edge research. Designed for physicists, chemists, materials scientists, and data scientists, this resource aims to enhance productivity and promote data-driven discoveries through accessible, standardized datasets.
Key Term Definitions:
| Name | Field/Area | Materials Types | Methods | Properties | Tools | Responsible Group | Institute/Affiliation | Total Entries | Active | Description |
|---|---|---|---|---|---|---|---|---|---|---|
| Materials science, AI, ML, and environmental engineering | Metal Organic Frameworks (MOFs), CO2, H2O adsorbates | DFT, High-Throughput DFT, ML | Electronic, Adsorption Energies, Structural, Thermodynamic Properties | ODAC23 Dataset, Leaderboards, ML Task Frameworks, | Fundamental AI Research (FAIR) at Meta and Researchers at Georgia Tech | Meta AI (FAIR) and Georgia Institute of Technology |
|
Yes | OpenDAC, a collaboration between Meta AI's FAIR and Georgia Tech, provides the ODAC23 dataset and ML tasks to accelerate AI-driven discovery of cost-effective sorbent materials for Direct Air Capture (DAC), advancing carbon dioxide removal technologies. | |
| Multidisciplinary, including science, social sciences, and humanities but highlighted here only about Materials science |
Nanomaterials, alloys, crystals, polymers, metal-organic frameworks (MOFs) |
DFT, MD, MBPT, DFPT, Transport, ML, XRD | Electronic, Mechanical, Thermodynamic, Adsorption Energies, Structural, Spectroscopic Properties | Web Interface, RESTful and OAI-PMH APIs, GitHub Integration | Zenodo team, supported by the OpenAIRE consortium and CERN's Digital Repositories Section | European Organization for Nuclear Research (CERN) , and OpenAIRE | 3 million documents | Yes | Zenodo, operated by CERN and OpenAIRE, is a free, open-access repository for sharing citable research outputs across all disciplines, assigning DOIs, supporting FAIR principles | |
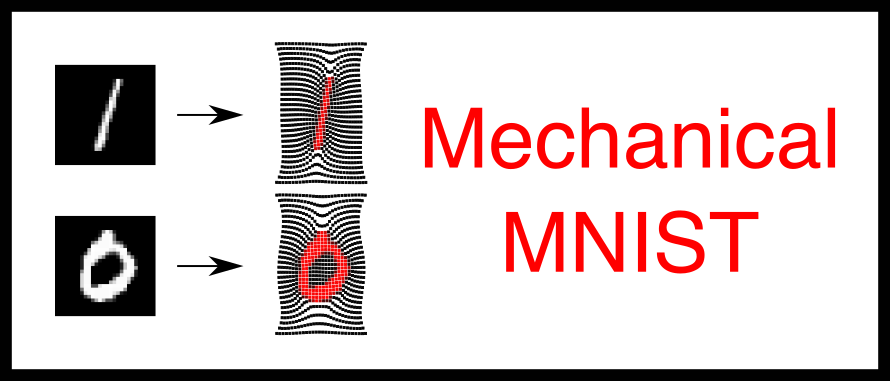
| Mechanical engineering, materials science, and computational mechanics | Neo-Hookean Materials, Heterogeneous Composites | Computer-aided design (CAD), Finite element methods (FEM), Bitmap-to-Material Mapping, ML | Mechanical properties, Dynamic properties (natural frequencies, damping ratios, modal masses) | FEniCS Scripts , Meta-Modeling Scripts, Visualization | Mechanical-MNIST Team and The Lejeune Lab, led by Prof. Emma Lejeune | Department of Mechanical Engineering, Boston University | 70000 + | Yes | Mechanical-MNIST is an open-access dataset of 70,000 FEniCS-based finite element simulations of heterogeneous Neo-Hookean materials derived from MNIST bitmaps, supporting meta-modeling for mechanical behavior under various load conditions. | |
| Materials science, additive manufacturing, metallurgy, and computational materials design | Open-Cell Metal Foams, Heterogeneous Cellular Structures | MD ML, X-ray Computed Tomography (XCT), Finite Element Analysis (FEA), Microstructural Reconstruction | Mechanical, Material Descriptors, Microstructural Properties | MDF’s faceted search, DREAM.3D | FOAM Development Team including Michael Groeber, Edwin Schwalbach | Air Force Research Laboratory (AFRL) , Argonne Leadership Computing Facility (ALCF) |
|
Yes | The FOAM Database is an open-source dataset hosted by the MDF at ALCF, providing 3D microstructural and mechanical property data for additively manufactured metallic foams to support materials science research and simulations. | |
| Materials science, quantum chemistry, computational chemistry, and machine learning | Crystalline solids, Metal-Organic Frameworks (MOFs), Organic and Inorganic Molecules, Polymers | DFT, MD, ML | Electronic, Thermodynamic, Adsorption Energies, Structural Properties | FAIRChem package (fairchem-core, fairchem-data-oc, fairchem-demo-ocpapi), OCPCalculator for ASE integration | Meta AI’s FAIR Chemistry team, including contributors like Muhammed Shuaibi | Meta AI’s Fundamental AI Research (FAIR) | 1000000 + | Yes | FAIR-Chem is an open-source repository of datasets, ML models, and workflows for advancing materials science and quantum chemistry, supporting simulations and discovery in catalysis and inorganic materials. It contains subdatasets like OC20 , OC22 , ODAC23 , and OMat24 and more | |
| Materials science, computational chemistry, inorganic crystal discovery, and machine learning | Inorganic Crystals, Transition Metal Compounds | DFT, MD, ML | Electronic, Thermodynamic, Optical Properties | Colab notebooks, Nequip (JAX implementation), github datasets | Google DeepMind Team including Amil Merchant and Ekin Dogus Cubuk | Google DeepMind | 520000 + | Yes | GNoME is an open-source dataset of inorganic crystal structures, including 381,000 novel stable materials, with DFT-calculated energies and properties, enabling large-scale materials discovery and simulations for applications like batteries and electronics. | |
| Materials Science, Machine Learning, Computational Materials Design, and Materials Informatics | Crystalline materials, Alloys, Perovskite, Metallic Glasses | DFT, ML | Electronic, Mechanical Properties | Formatted for MAST-ML (Materials Simulation Toolkit - Machine Learning), github | Ryan Jacobs, Daniel Sauceda, and James Cumby | University of Wisconsin-Madison ( Department of Materials Science and Engineering ) and University of Edinburgh |
|
Yes | MAST-ML Education Datasets are open-source collections of materials science data (dilute solute diffusion, perovskite stability, and metallic glasses) designed for machine learning education and property prediction using the MAST-ML toolkit. | |
| Computational Quantum Chemistry, Materials Science, Drug Discovery, and Machine Learning |
Organic and inorganic molecules including drugs, toxins
|
ML, Molecular Featurization
|
Molecular Properties | HuggingFaceFeaturizer , SmilesTokenizer , ChemBERTa , github DeepChem | DeepChem community including Bharath Ramsundar, Peter Eastman, Patrick Walters | Initially Stanford University, now a community-driven project | 1000000 + | Yes | DeepChem is an open-source repository of datasets and ML models for computational chemistry and drug discovery, enabling molecular property prediction and simulations with tools like SmilesTokenizer and HuggingFaceFeaturizer. | |
| Materials Science, AI, ML, electronic structure, force-field simulations, quantum computation, and experimental materials research | Crystalline solids, Metal-Organic Frameworks (MOFs), Organic and inorganic molecules |
DFT, Classical Force Fields (FF), ML
|
Electronic, Adsorption, Mechanical, Structural, Thermodynamic Properties | GitHub Actions for automated testing, mkdocs for visualization, JARVIS-Tools | Kamal Choudhary, Daniel Wines, Kevin Garrity, Aldo Romero, Jaron Krogel, Kayahan Saritas, Panchapakesan Ganesh | National Institute of Standards and Technology (NIST), USA | 80000 + | Yes | JARVIS-Leaderboard is an open-source platform for benchmarking materials science methods (AI, ML, DFT, force-fields, quantum computation, experiments) using JARVIS-Tools datasets to ensure reproducibility and transparency in materials design. | |
| Materials science, computational chemistry, machine learning, and inorganic materials discovery | Crystalline Solids, Materials Project Structures | DFT, MD, ML | Electronic, Mechanical , Thermodynamic, Optical Properties | OPTIMADE API, M3GNet , web interface , materialsvirtuallab | Chi Chen, Shyue Ping Ong, and the Materials Virtual Lab team | University of California San Diego ( Jacobs School of Engineering ) and Materials Virtual Lab | 180000 + | Yes | Matterverse.ai developed by the Materials Virtual Lab at UC San Diego, is a database (not completely free) of over 31 million hypothetical materials with M3GNet-predicted properties, facilitating AI-driven discovery of novel materials for technological applications. | |
| Computational Quantum Chemistry, and Quantum Machine Learning for molecular property prediction | Organic and Organometallic Compounds, COD structures | DFT, DFPT, ML | Electronic, Geometric, Thermodynamic, Spectroscopic Properties | sGDML , SchNetPack | QMO Team including O. Anatole von Lilienfeld, Klaus-Robert Müller, Alexandre Tkatchenko, Matthias Rupp, and others | Various institutions, including University of Basel , TU Berlin , and Fritz Haber Institute | 7000 + | Yes | Quantum-Machine.org is an open-source repository of datasets (e.g., QM7 , QM9 ) and software (e.g., sGDML, SchNetPack) for accelerating quantum machine learning and quantum chemistry simulations. | |
| Molecular machine learning, computational chemistry, drug discovery, materials science, biophysics | Organic Molecules, Organometallic Compounds, Biomolecules, Drug-like Molecules, Organic photovoltaic materials | DFT, MD, ML, Data Splitting and Benchmarking | Electronic, Chemical, Biophysical, Physiological Properties | DeepChem library , MolGraphConvFeaturizer , dataset loaders (load_qm9, load_tox21) | DeepChem community, including Zhenqin Wu, Bharath Ramsundar, Evan N. Feinberg, Joseph Gomes, and others | Initially Stanford University, now a community-driven project with contributions from various institutions | 700000 + | Yes | MoleculeNet, part of the DeepChem library, is an open-source benchmarking framework for molecular machine learning, providing curated datasets, evaluation metrics, and ML algorithms for predicting molecular properties in chemistry and drug discovery. | |
| Computational quantum chemistry, molecular machine learning, drug discovery, and chemical science | Organic Molecules, Drug-like Molecules | DFT, ML |
Electronic, Structural, Molecular Properties, Conformational Data
|
HamiltonianDatabase and ASENablaDFT, model_registry | nablaDFT Team including Kuzma Khrabrov, Ilya Shenbin, Alexander Ryabov, Artem Tsypin | Artificial Intelligence Research Institute (AIRI), Russia | 190000 + | Yes | nablaDFT is an open-source dataset and benchmark containing DFT-calculated properties, designed for training and evaluating neural network potentials in quantum chemistry and drug discovery. | |
| Computational Chemistry, Molecular ML, drug discovery, and protein-ligand interactions | Organic Molecules, Drug-like Molecules, Biomolecules, Organic-Inorganic Hybrids, Water Cluster | DFT, MD, ML | Electronic, Structural, Chemical, Interaction Properties | OpenFF-QCSubmit, QCFractal, Psi4 for QM calculations, createSpiceDataset.py for TorchMD-Net compatibility | SPICE Team including Peter Eastman, Pavan Kumar Behara, David L. Dotson, Raimondas Galvelis, John E. Herr | OpenMM, OpenFF, MolSSI, and collaborating institutions like Stanford University and Memorial Sloan Kettering Cancer Center | 110000 + | Yes |
SPICE, developed by the OpenMM team, is an open-source dataset of over 1.1 million quantum mechanical conformations for drug-like molecules and peptides, designed for training machine learning potentials in molecular simulations.
|
|
| Cheminformatics, Computational Quantum Chemistry, Materials Science, and AI for scientific discovery | Organic Molecules, Inorganic Materials, Biomolecules, Organic-Inorganic Hybrids | DFT, MD, ML | Electronic, Structural, Thermodynamic, Physical, Biological, Properties | PotNet, ComENet, GraphBP, SineNet, LatentDiff, QH9 Dataset/Benchmark | DIVE (Data Integration, Visualization, and Exploration) Lab Development Team | DIVE Lab , Texas A&M University , Department of Computer Science and Engineering | 134000 + | Yes |
AIRS is an open-source and aids AI-driven scientific discovery in quantum and continuum systems. It handles tasks like molecular property modeling with tools like PotNet and ComENet, integrating with PyTorch Geometric and JARVIS for researchers in materials science and computational physics. |